In a groundbreaking study published in Nature Communications, researchers from Vanderbilt University and UC San Diego have created the first high-resolution metabolic map showing how glucose is processed inside cells—from whole animals down to individual organelles. This multi-scale approach combines stable isotope tracing, advanced microscopy, and AI-powered image analysis to reveal the spatial organization of glucose metabolism in unprecedented detail.
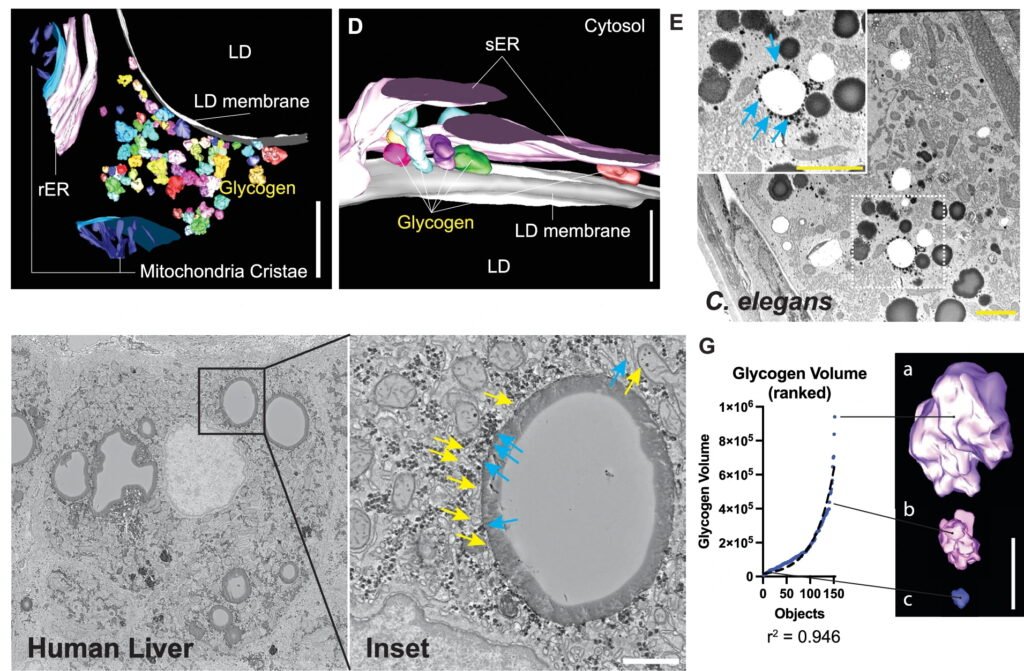
Led by Rafael Arrojo e Drigo, the team infused live mice with isotopically labeled glucose and tracked how glucose-derived carbons were incorporated into cellular structures like glycogen, lipid droplets, and mitochondria. The researchers discovered a previously unknown interaction between lipid droplets and glycogen synthesis, suggesting that these organelles work together to regulate energy storage and usage. They also observed dynamic shifts in contact points between mitochondria and the endoplasmic reticulum—key organelles involved in energy production and nutrient sensing—depending on blood glucose levels.
This spatial reorganization forms part of a broader organelle network that coordinates metabolic responses within the cell. By charting these interactions over time, the study sheds light on how cells adapt to changing nutrient conditions and maintain energy balance. The findings have implications for understanding metabolic diseases like diabetes, obesity, and cancer, as well as aging and neurodegeneration.
Traditional metabolomics methods analyze bulk tissue samples without accounting for spatial context. This new technique bridges that gap, offering a window into the subcellular architecture of metabolism. The Vanderbilt team collaborated with experts in mass spectrometry, computational modeling, and microscopy from the National Center for Microscopy and Imaging Research (NCMIR) at UCSD, creating a pipeline that spans animal physiology, tissue biology, and molecular dynamics.
The study also highlights the importance of interdisciplinary collaboration. Co-first authors Christopher Acree and Aliyah Habashy worked alongside scientists from Vanderbilt’s Mouse Metabolic Phenotyping Center, the Mass Spectrometry Research Center, and the Department of Cell and Developmental Biology. Their work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute on Aging, and the Glenn Foundation for Medical Research.
Looking ahead, the team hopes to apply this method to other tissues and disease models, exploring how spatial nutrient organization influences health and pathology. By visualizing glucose metabolism at the single-cell level, they’ve opened a new frontier in metabolic research—one that could lead to more precise diagnostics, targeted therapies, and a deeper understanding of how the body manages its most essential fuel.
Article from Vanderbilt University: Pioneering new method reveals glucose channeling, charting the fine structure of energy metabolism inside active cells
Abstract in Nature Communications: Spatial patterns of hepatocyte glucose flux revealed by stable isotope tracing and multi-scale microscopy